Display 2D tour projections displayed separately by groups
Source:R/display-groupxy.r
display_groupxy.RdThis function is designed to allow comparisons across multiple groups, especially for examining things like two (or more) different models on the same data. The primary display is a scatterplot, with lines or contours overlaid.
display_groupxy(
centr = TRUE,
axes = "center",
half_range = NULL,
col = "black",
pch = 20,
cex = 1,
edges = NULL,
edges.col = "black",
edges.width = 1,
group_by = NULL,
plot_xgp = TRUE,
palette = "Zissou 1",
shapeset = c(15:17, 23:25),
axislablong = FALSE,
...
)
animate_groupxy(data, tour_path = grand_tour(), ...)Arguments
- centr
if TRUE, centers projected data to (0,0). This pins the center of data cloud and make it easier to focus on the changing shape rather than position.
- axes
position of the axes: center, bottomleft or off
- half_range
half range to use when calculating limits of projected. If not set, defaults to maximum distance from origin to each row of data.
- col
color to use for points, can be a vector or hexcolors or a factor. Defaults to "black".
- pch
shape of the point to be plotted. Defaults to 20.
- cex
size of the point to be plotted. Defaults to 1.
- edges
A two column integer matrix giving indices of ends of lines.
- edges.col
colour of edges to be plotted, Defaults to "black"
- edges.width
line width for edges, default 1
- group_by
variable to group by. Must have less than 25 unique values.
- plot_xgp
if TRUE, plots points from other groups in light grey
- palette
name of color palette for point colour, used by
hcl.colors, default "Zissou 1"- shapeset
numbers corresponding to shapes in base R points, to use for mapping categorical variable to shapes, default=c(15:17, 23:25)
- axislablong
text labels only for the long axes in a projection, default FALSE
- ...
other arguments passed on to
animateanddisplay_groupxy- data
matrix, or data frame containing numeric columns
- tour_path
tour path generator, defaults to 2d grand tour
Examples
animate_groupxy(flea[, 1:6], col = flea$species,
pch = flea$species, group_by = flea$species)
#> Converting input data to the required matrix format.
#> Using half_range 4.4
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
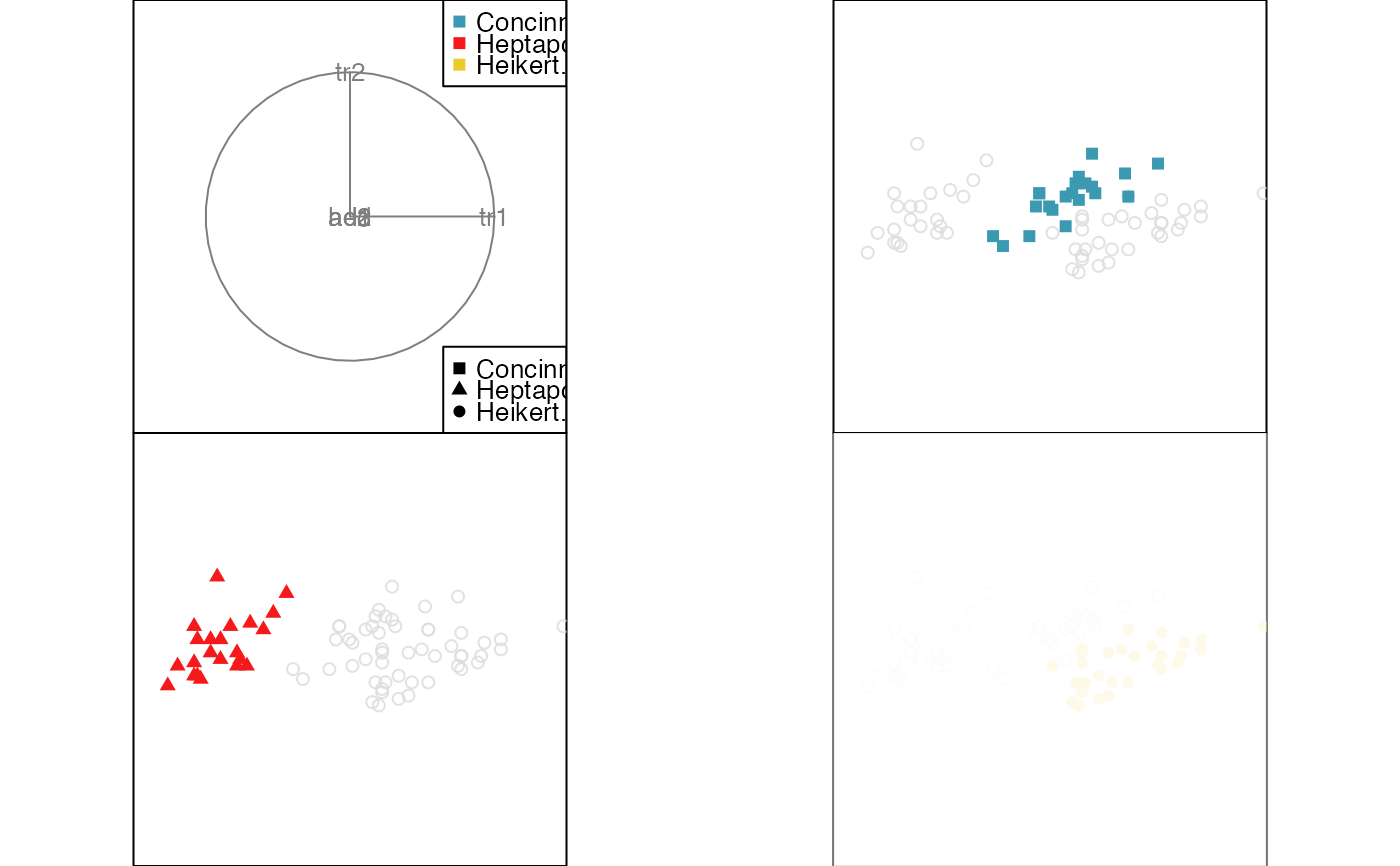 #> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
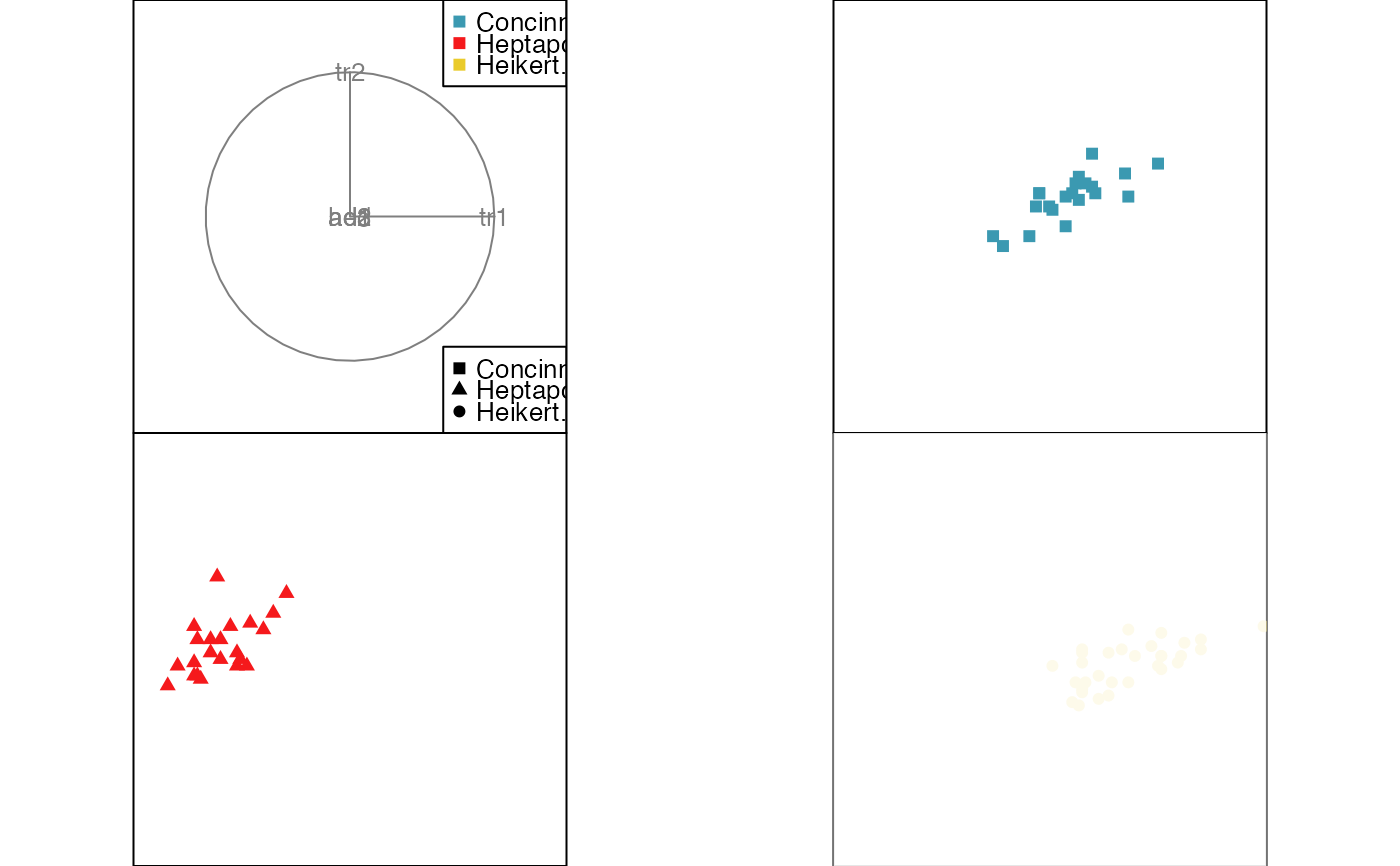
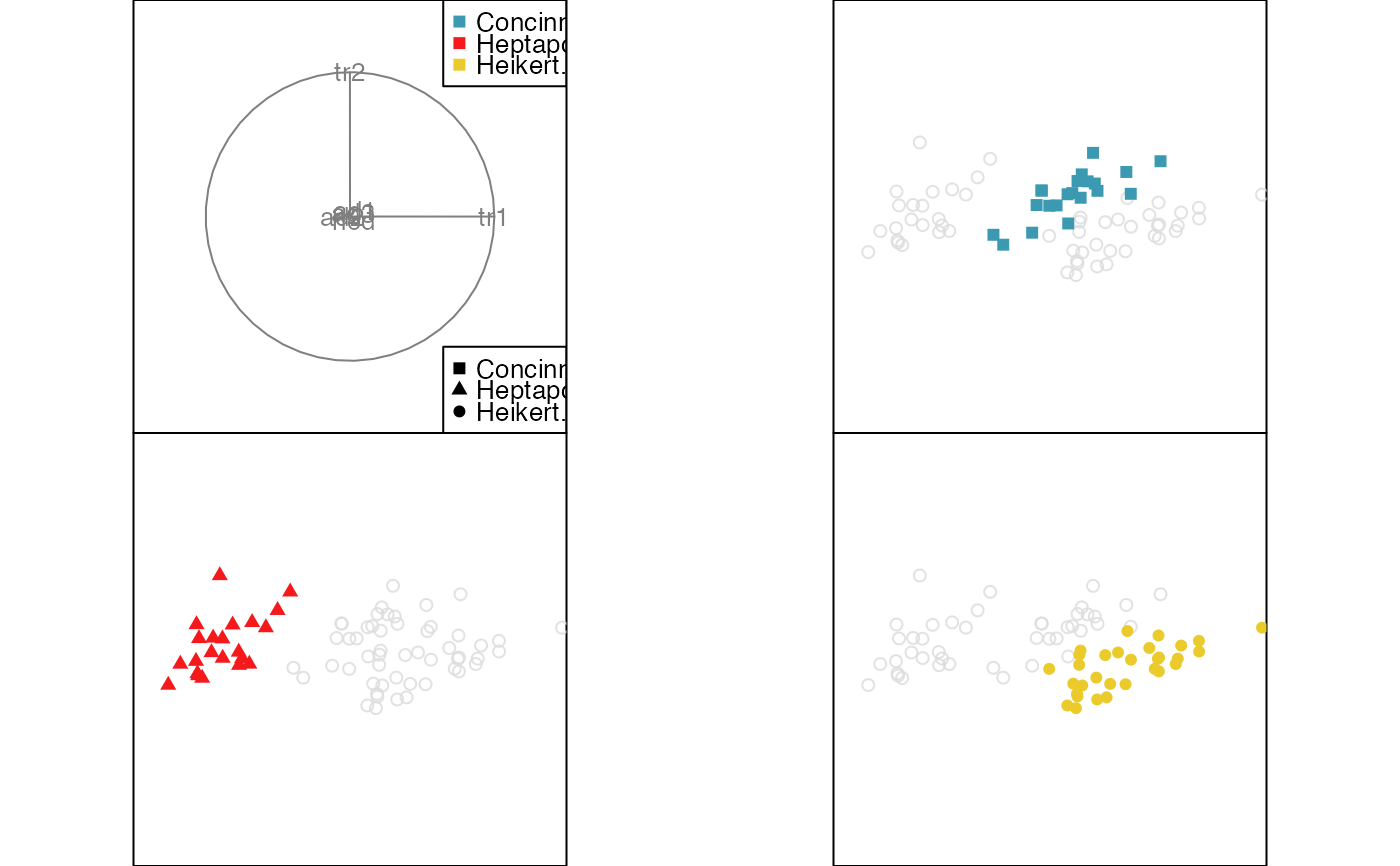 animate_groupxy(flea[, 1:6], col = flea$species,
pch = flea$species, group_by = flea$species,
plot_xgp = FALSE)
#> Converting input data to the required matrix format.
#> Using half_range 4.4
animate_groupxy(flea[, 1:6], col = flea$species,
pch = flea$species, group_by = flea$species,
plot_xgp = FALSE)
#> Converting input data to the required matrix format.
#> Using half_range 4.4
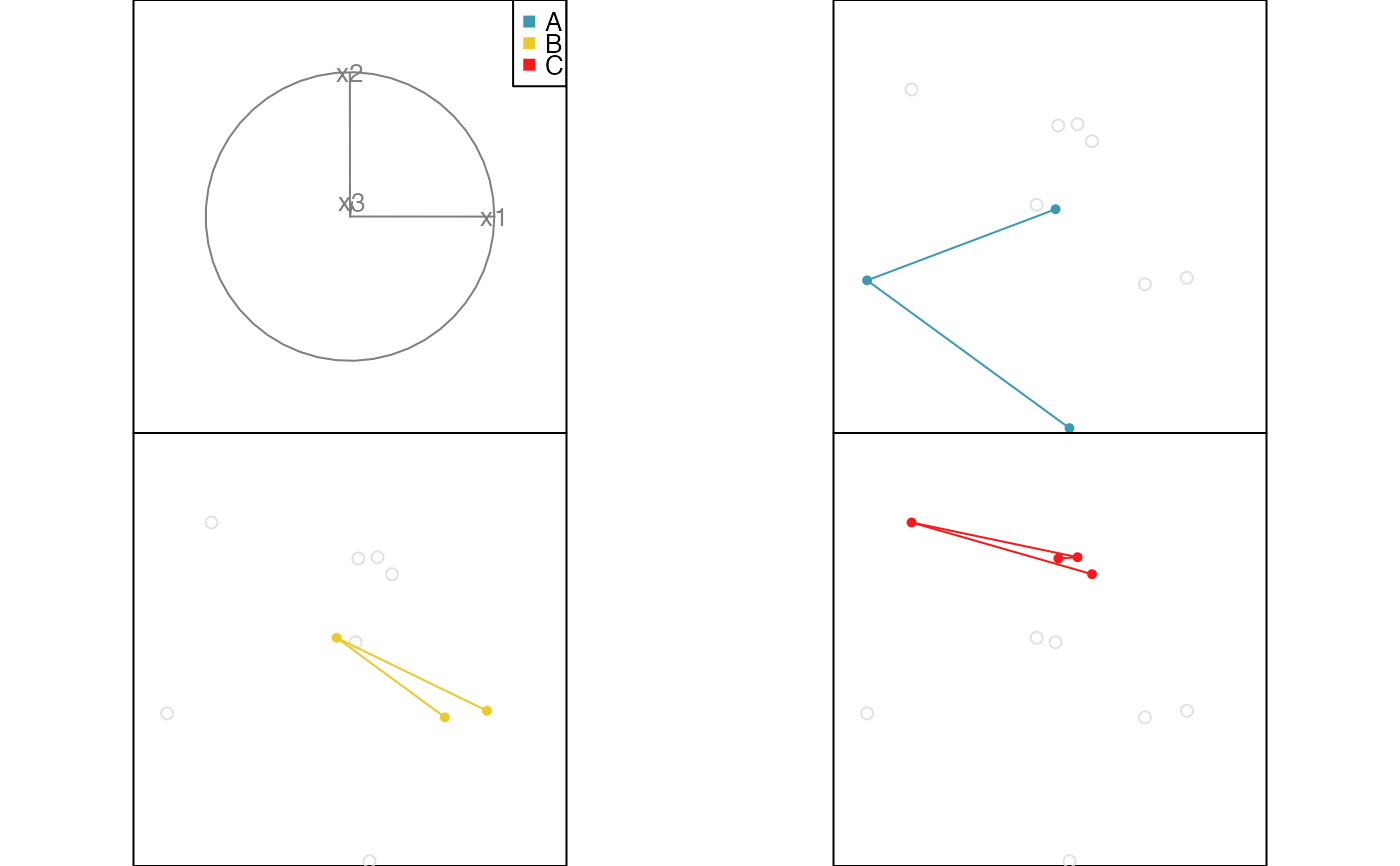
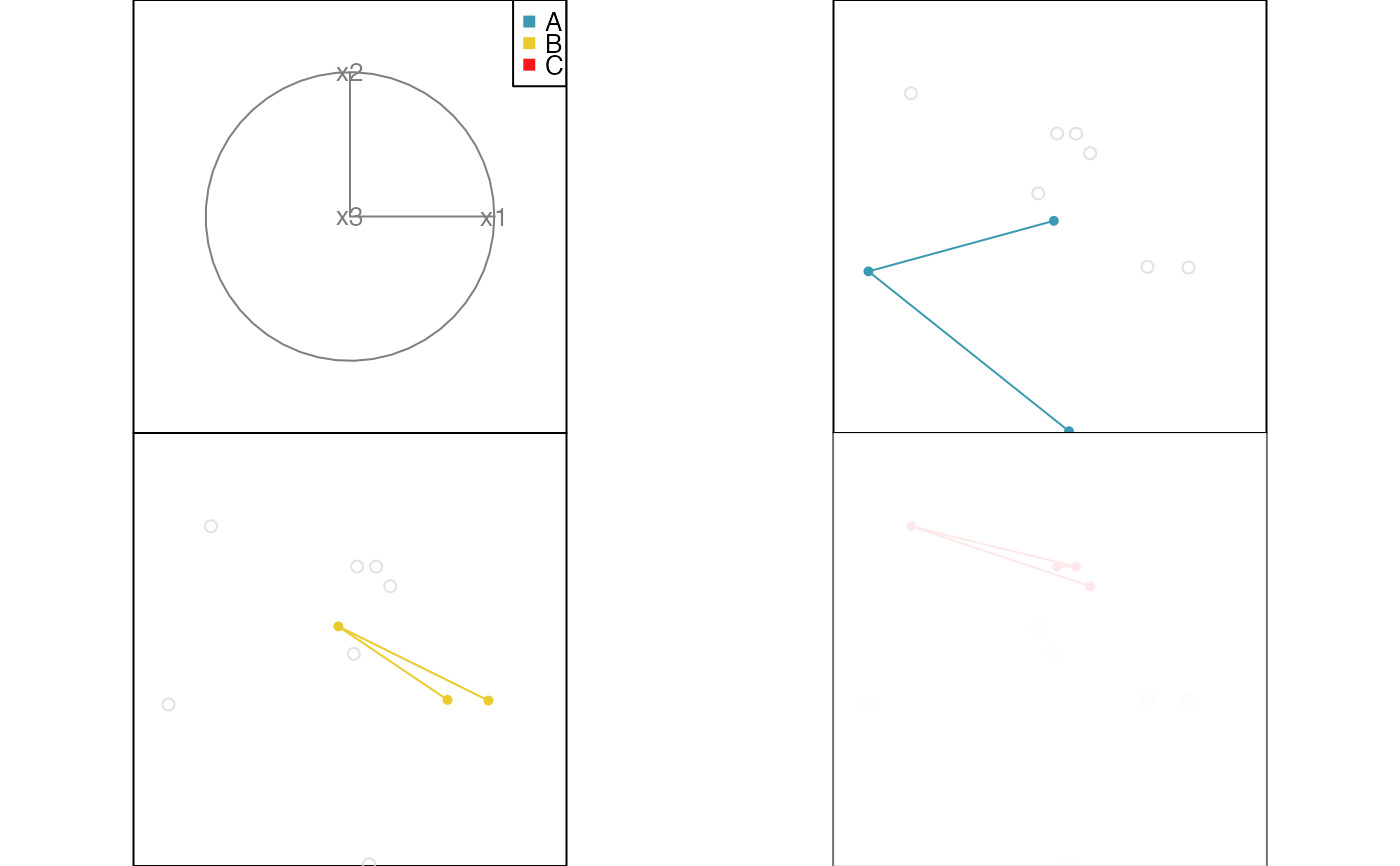
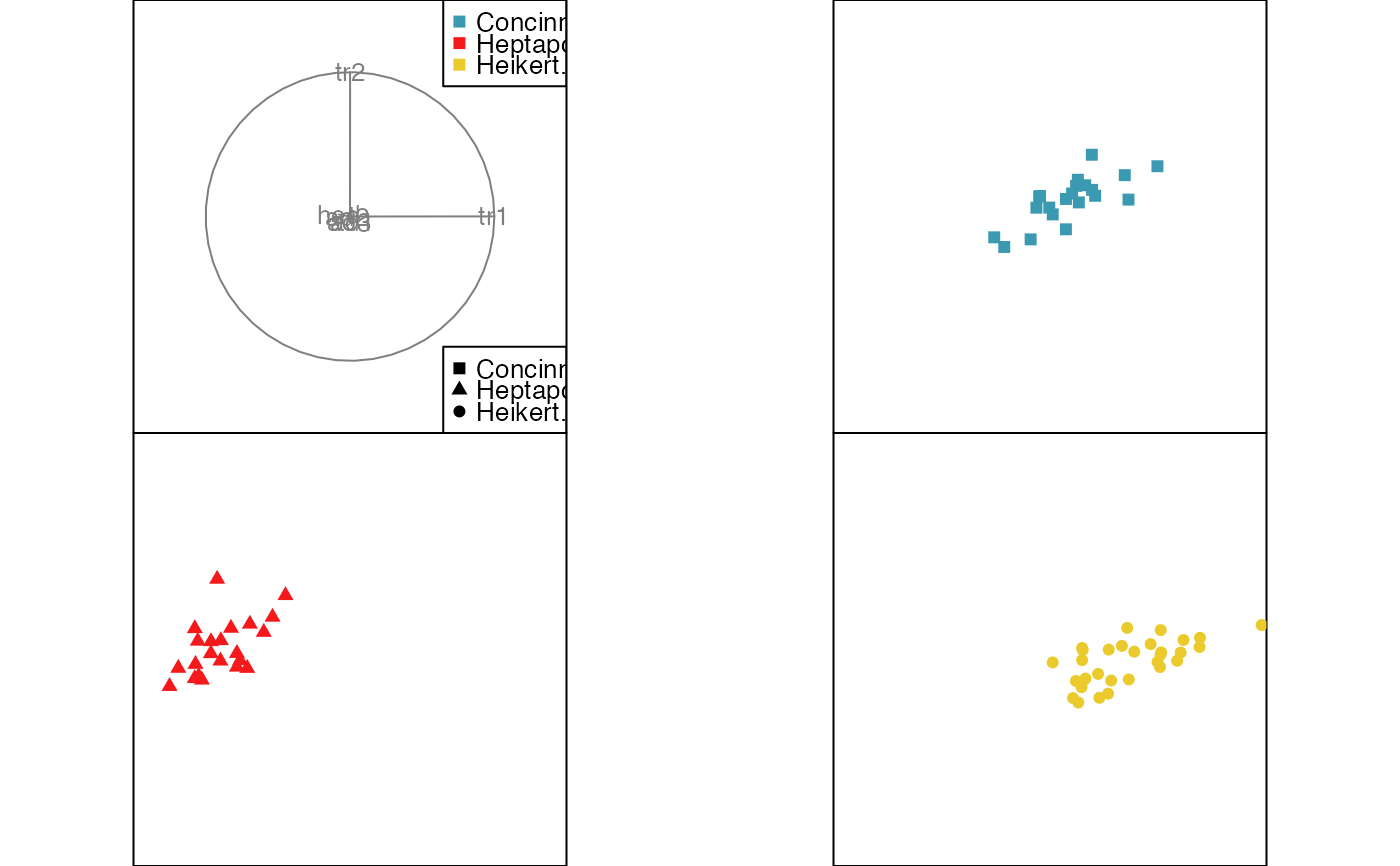 # Edges example
x <- data.frame(x1=runif(10, -1, 1), x2=runif(10, -1, 1), x3=runif(10, -1, 1))
x$cl <- factor(c(rep("A", 3), rep("B", 3), rep("C", 4)))
x.edges <- cbind(from=c(1,2, 4,5, 7,8,9), to=c(2,3, 5,6, 8,9,10))
x.edges.col <- factor(c(rep("A", 2), rep("B", 2), rep("C", 3)))
animate_groupxy(x[,1:3], col=x$cl, group_by=x$cl, edges=x.edges, edges.col=x.edges.col)
#> Converting input data to the required matrix format.
#> Using half_range 1.3
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
# Edges example
x <- data.frame(x1=runif(10, -1, 1), x2=runif(10, -1, 1), x3=runif(10, -1, 1))
x$cl <- factor(c(rep("A", 3), rep("B", 3), rep("C", 4)))
x.edges <- cbind(from=c(1,2, 4,5, 7,8,9), to=c(2,3, 5,6, 8,9,10))
x.edges.col <- factor(c(rep("A", 2), rep("B", 2), rep("C", 3)))
animate_groupxy(x[,1:3], col=x$cl, group_by=x$cl, edges=x.edges, edges.col=x.edges.col)
#> Converting input data to the required matrix format.
#> Using half_range 1.3
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
 #> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter
#> Warning: "shapes" is not a graphical parameter